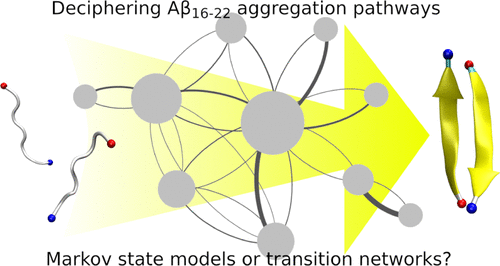
Molecular dynamic (MD) simulations are an important tool for studying protein aggregation processes, which play a central role in a number of diseases including Alzheimer’s disease. However, MD simulations produce large amounts of data, requiring advanced methods to extract mechanistic insight into the process under study. Transition networks (TNs) provide an elegant method to identify (meta)stable states and the transitions between them from MD simulations. Here, we apply two different methods to generate TNs for protein aggregation: Markov state models (MSMs), which are based on kinetic clustering the state space, and TNs using conformational clustering.
Alexander-Maurice Illig & Birgit Strodel